Chapter 5 Data Transformation 3: Tidy Data
Intended Learning Outcomes
The whole purpose of the next two lecture is to tidy up dataframe and expose you to a bunch of useful functions that would make your daily life easier when dealing with your own data. By the end of today, you will know
- How to rearrange data from long format into to wide format and vice versa
- How to combine and separate columns
This lesson is led by Gaby Mahrholz.
5.1 Pre-steps
All the functions for tidying data are within tidyverse so we need to make sure to load it into the library. Today’s lecture will make use of built-in dataframes which means we do not have to load any data into our Global Environment for the minute.
library(tidyverse)5.2 Tidy data
“Happy families are all alike; every unhappy family is unhappy in its own way.” – Leo Tolstoy
“Tidy datasets are all alike, but every messy dataset is messy in its own way.” – Hadley Wickham
In this chapter, you will learn a consistent way to organise your data in R. Hadley Wickham and Garrett Grolemund call this tidy data. In most cases, data does not come in a tidy format. Researchers spend most of their time getting the data into the shape it needs to be for a particular analysis. The tools we introduce you to this week and next should form the basics for data wrangling. The idea is that tidy datasets are easier to work with in the long run.
There are three interrelated rules which make a dataset tidy:
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
If you want to know more about tidy data, Hadley Wickham’s paper can be found on https://vita.had.co.nz/papers/tidy-data.pdf or check out chapter 12 in the book R for Data Science (https://r4ds.had.co.nz/tidy-data.html).
5.3 gather() and spread()
gather() and spread() are part of tidyr (also part of the tidyverse package) and help you to rearrange the shape of your data. To get your data into long format, you would use gather(); if you wanted to get your data from long to wide format, you would need spread(). Here is a nice gif animation by Garrick Aden-Buie (https://www.garrickadenbuie.com/project/tidyexplain/#spread-and-gather) that shows you how gather() and spread() work.
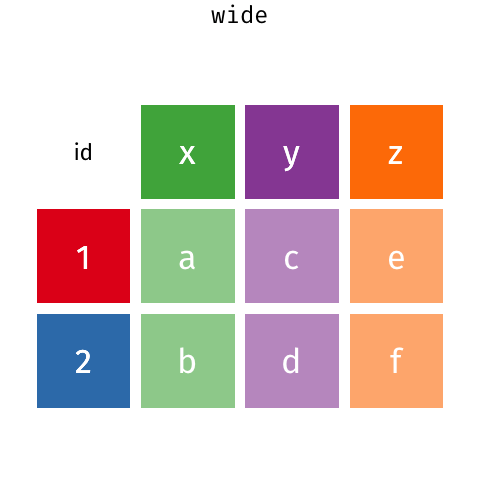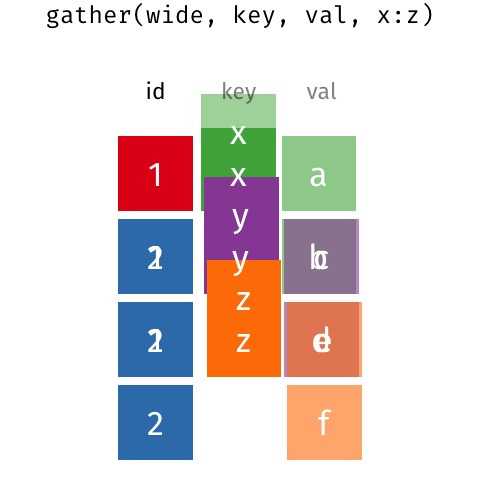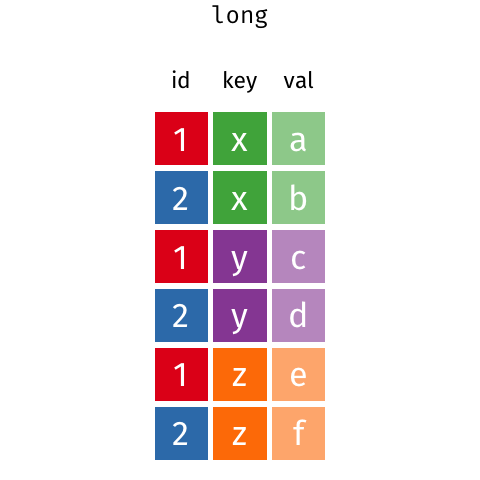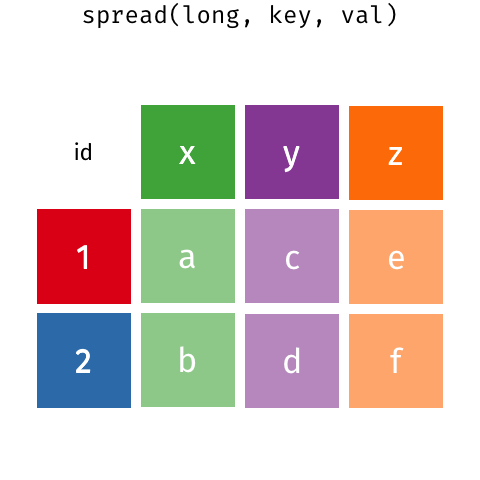
5.3.1 gather()
We can start this lecture by looking at a practical example in the build-in dataset table1 to table5. They show the number of Tuberculosis cases documented by the World Health Organization in Afghanistan, Brazil, and China between 1999 and 2000 (the full dataset is called who). The data is part of the tidyverse package, so as long as we have tidyverse loaded into the library, there should not be an issue in accessing this dataset.
table1## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583table1 has a very tidy structure as it follows the three principles above - each variable is its own column, each observation is its own row, and each value is its own cell.

You can see the data contains values associated with four variables (country, year, cases, and population). table4a however, looks a touch not right.
table4a## # A tibble: 3 x 3
## country `1999` `2000`
## * <chr> <int> <int>
## 1 Afghanistan 745 2666
## 2 Brazil 37737 80488
## 3 China 212258 213766
Somehow the values for the columns year and cases are missing and instead we find two separate columns for 1999 and 2000 that have all cases values listed. This is a problem as 1999 and 2000 are actual values and not variables. What we want to do, is gather 1999 and 2000 in one column year and respective values into a second column called cases.
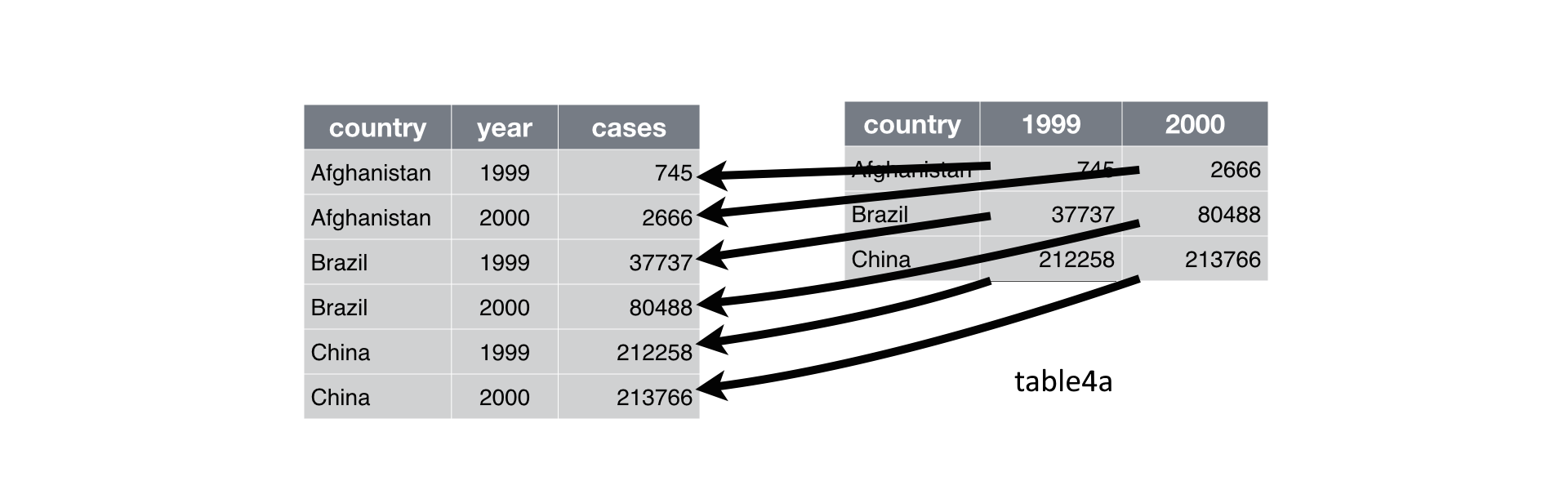
Currently the data is in what is called a wide format. The function gather() comes in handy when we want to get the data into the shape of a long format like in table1. gather() is structured like:
Let’s apply this formula to table4a
- We need a dataframe that we want to modify ->
table4a - We need to come up with the name of a new column in which the values of 1999 and 2000 will be stored ->
year - We need to come up with the name of a new column in which all the case values will be stored ->
cases - And finally, we need to tell R which columns in
table4awe want to turn into long format (i.e. the columns of the 7 subscales) -> 1999 and 2000 (Note that “1999” and “2000” are non-syntactic names as they don’t start with a letter. The solution is to surround them in backticks.)
gather(table4a, key = year, value = cases, `1999`:`2000`)## # A tibble: 6 x 3
## country year cases
## <chr> <chr> <int>
## 1 Afghanistan 1999 745
## 2 Brazil 1999 37737
## 3 China 1999 212258
## 4 Afghanistan 2000 2666
## 5 Brazil 2000 80488
## 6 China 2000 213766
Here we have only 2 columns to reshape 1999 and 2000. We could have written gather(table4a, year, cases, `1999`, `2000`).
Your turn
Look at table4b. There are two separate columns 1999 and 2000 with population values for Afghanistan, Brazil, and China. Use the gather() function on table4b to reshape it from wide format into long format.
gather(table4b, key = year, value = population, `1999`:`2000`)## # A tibble: 6 x 3
## country year population
## <chr> <chr> <int>
## 1 Afghanistan 1999 19987071
## 2 Brazil 1999 172006362
## 3 China 1999 1272915272
## 4 Afghanistan 2000 20595360
## 5 Brazil 2000 174504898
## 6 China 2000 12804285835.3.2 spread()
Now onto the reverse. spread() will spread the data from long into wide format. When you have observations that are scattered across multiple rows, you can use spread() to split those rows up into separate variables. Let’s take a look at table2.
table2## # A tibble: 12 x 4
## country year type count
## <chr> <int> <chr> <int>
## 1 Afghanistan 1999 cases 745
## 2 Afghanistan 1999 population 19987071
## 3 Afghanistan 2000 cases 2666
## 4 Afghanistan 2000 population 20595360
## 5 Brazil 1999 cases 37737
## 6 Brazil 1999 population 172006362
## 7 Brazil 2000 cases 80488
## 8 Brazil 2000 population 174504898
## 9 China 1999 cases 212258
## 10 China 1999 population 1272915272
## 11 China 2000 cases 213766
## 12 China 2000 population 1280428583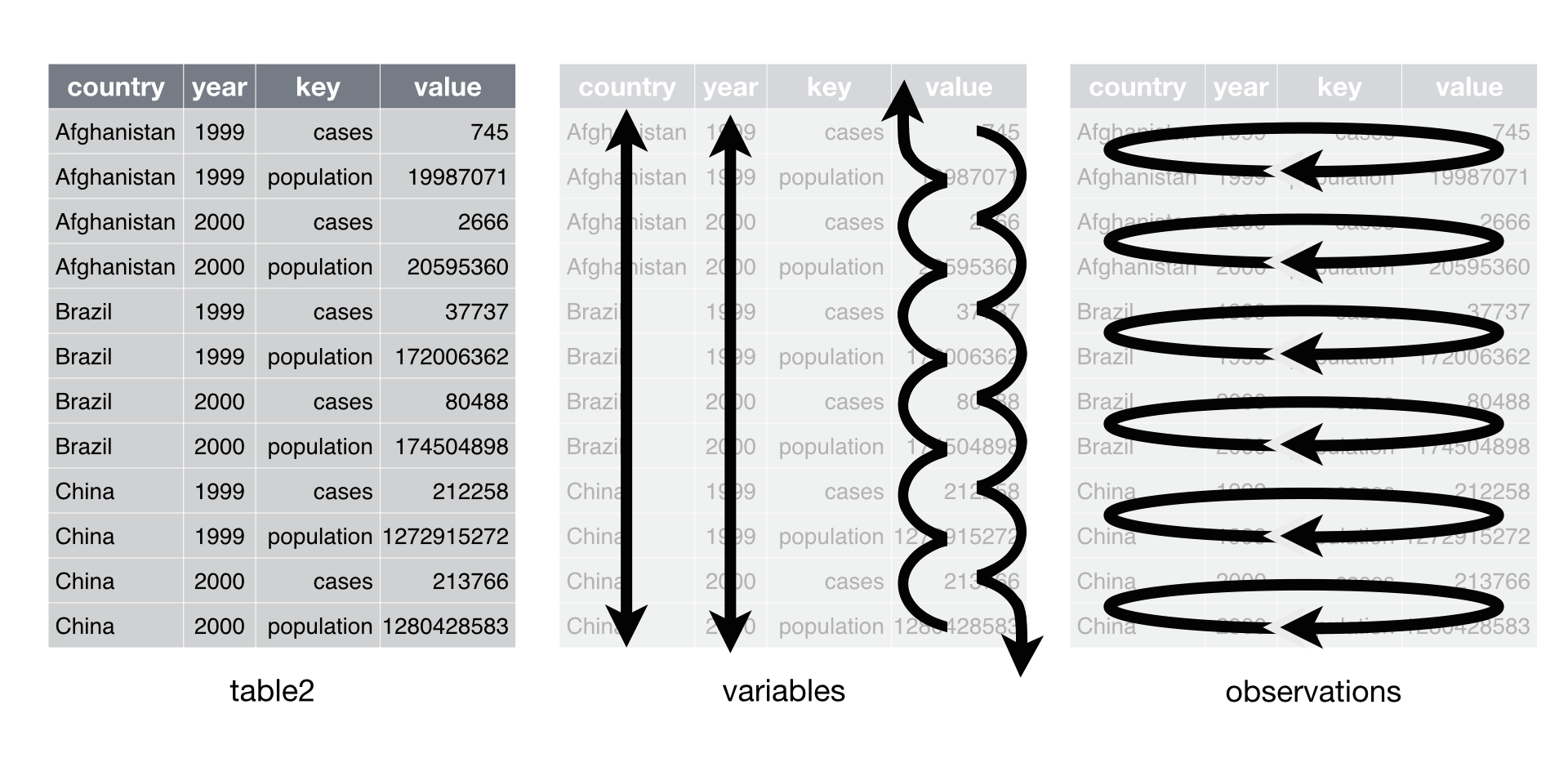
table2 has the variables cases and population stored in one column type and their respective values in count. However, cases and population are separate variables rather than values of type. So best to get them spread out into their own columns as shown in the image below.
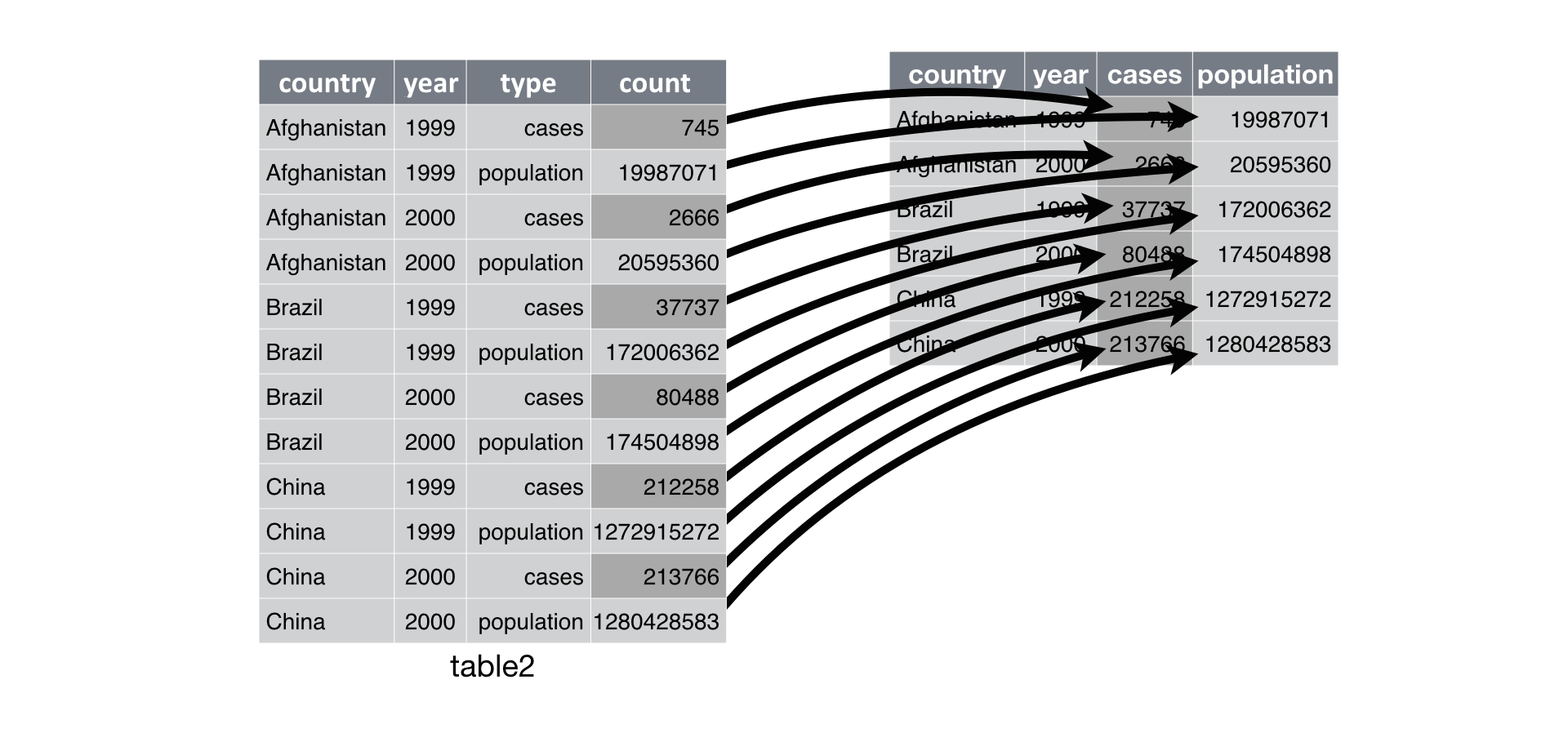
Take a look how spread() achieves that.
Let’s apply the formula above to table2:
- We need a dataframe that we want to modify ->
table2 - We need to decide which of the columns in
table2will become the new column headers ->type - We need to decide which of the columns in
table2has all the data in it that will become the new cell values ->count
spread(table2, key = type, value = count)## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583Voila, cases and population are in their own columns.
Let me modify table1 a tiny bit.
table1_mod <- table1 %>%
mutate(percent = cases/ population * 100) %>%
gather(variable, values, cases:percent)| country | year | variable | values |
|---|---|---|---|
| Afghanistan | 1999 | cases | 7.450000e+02 |
| Afghanistan | 2000 | cases | 2.666000e+03 |
| Brazil | 1999 | cases | 3.773700e+04 |
| Brazil | 2000 | cases | 8.048800e+04 |
| China | 1999 | cases | 2.122580e+05 |
| China | 2000 | cases | 2.137660e+05 |
| Afghanistan | 1999 | population | 1.998707e+07 |
| Afghanistan | 2000 | population | 2.059536e+07 |
| Brazil | 1999 | population | 1.720064e+08 |
| Brazil | 2000 | population | 1.745049e+08 |
| China | 1999 | population | 1.272915e+09 |
| China | 2000 | population | 1.280429e+09 |
| Afghanistan | 1999 | percent | 3.727400e-03 |
| Afghanistan | 2000 | percent | 1.294470e-02 |
| Brazil | 1999 | percent | 2.193930e-02 |
| Brazil | 2000 | percent | 4.612360e-02 |
| China | 1999 | percent | 1.667500e-02 |
| China | 2000 | percent | 1.669490e-02 |
Your turn
In the modified table above table1_mod, we added a percent column to see how much percentage of the population had Tuberculosis in 1999 and 2000 in each of the countries Afghanistan, Brazil, and China. But somehow, all numerical information has ended up in 1 column (values).
Your task is to spread the data back out, so that cases, population, and percentage values are in their own columns, titled as cases, population, and percent respectively. Store the new object in the Global Environment as table1_mod_corrected. When you are done, table1_mod_corrected should look like this…
| country | year | cases | percent | population |
|---|---|---|---|---|
| Afghanistan | 1999 | 745 | 0.0037274 | 19987071 |
| Afghanistan | 2000 | 2666 | 0.0129447 | 20595360 |
| Brazil | 1999 | 37737 | 0.0219393 | 172006362 |
| Brazil | 2000 | 80488 | 0.0461236 | 174504898 |
| China | 1999 | 212258 | 0.0166750 | 1272915272 |
| China | 2000 | 213766 | 0.0166949 | 1280428583 |
table1_mod_corrected <- spread(table1_mod, key = variable, value = values)5.4 separate() and unite()
separate() turns a single character column into multiple columns, whereas unite() combines the values of multiple columns into one column.
5.4.1 separate()
Take a look at table3.
table3## # A tibble: 6 x 3
## country year rate
## * <chr> <int> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583In table3, two out of three principles of tidy data are violated. There are multiple values stored in column rate (rule 3) but these multiple values also belong to two different variables - cases and population respectively (rule 1).
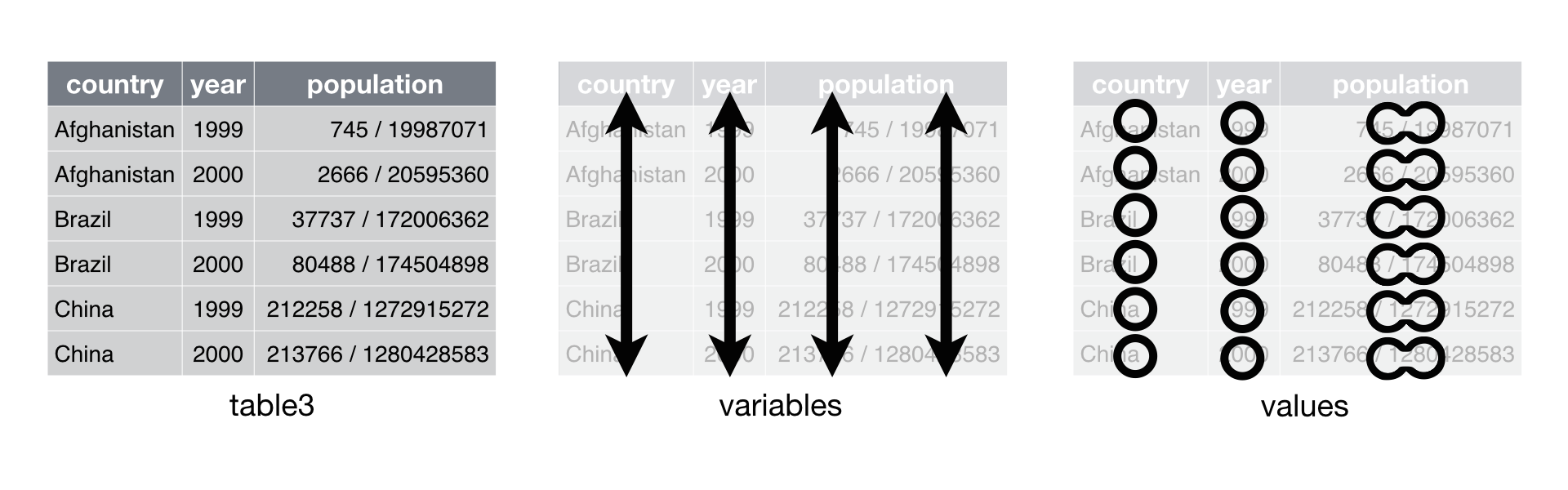
Here, we would use separate() to split the column rate into two columns cases and population.

The function separate() is structured like this:
We would need
- the data in which the column to separate is located ->
table3, - the actual column we want to separate ->
rate, - the into argument asking into how many columns we want to separate the old column and what we want to call these new columns ->
c("cases", "population"), and - a separator argument ->
"/".
separate(table3, rate, into = c("cases", "population"), sep = "/")## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <chr> <chr>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583R has put all cell information before the / into a column we named cases, and everything after the / into the column we decided to call population. / will not appear in either column cases or population. The data is now in a tidy format.
By the way, the separator argument sep = can be any non-alphanumeric character, for example:
- underscore (“_“),
- comma (“,”),
- semi-colon (“;”),
- colon (“:”),
- forward-slash (“/”),
- space (" “) or
- nothing (“”).
But watch out! When the separator is a full stop, you would need to add a double backward slash in front of it (sep = “\\.”).
5.4.2 additional arguments for separate()
5.4.2.1 Retaining the original column
If for some reason, you wanted to keep the original column, you can set an additional argument remove to FALSE.
separate(table3, rate, into = c("cases", "population"), sep = "/", remove = FALSE)## # A tibble: 6 x 5
## country year rate cases population
## <chr> <int> <chr> <chr> <chr>
## 1 Afghanistan 1999 745/19987071 745 19987071
## 2 Afghanistan 2000 2666/20595360 2666 20595360
## 3 Brazil 1999 37737/172006362 37737 172006362
## 4 Brazil 2000 80488/174504898 80488 174504898
## 5 China 1999 212258/1272915272 212258 1272915272
## 6 China 2000 213766/1280428583 213766 12804285835.4.2.2 Dropping parts of the original column
What if we were just interested retaining part of the data in the cell, for example keeping the column cases but not population. The solution would be to work with an NA argument. Defining the new columns as cases and NA will keep everything before the separator in the column cases, and drops everything after the separator.
separate(table3, rate, into = c("cases", NA), sep = "/")## # A tibble: 6 x 3
## country year cases
## <chr> <int> <chr>
## 1 Afghanistan 1999 745
## 2 Afghanistan 2000 2666
## 3 Brazil 1999 37737
## 4 Brazil 2000 80488
## 5 China 1999 212258
## 6 China 2000 213766Your turn
Separate the column rate in table3 to keep only the values after the separator in a column called population. Drop the values in front of the separator. Retain the column rate as well.
separate(table3, rate, into = c(NA, "population"), sep = "/", remove = FALSE)## # A tibble: 6 x 4
## country year rate population
## <chr> <int> <chr> <chr>
## 1 Afghanistan 1999 745/19987071 19987071
## 2 Afghanistan 2000 2666/20595360 20595360
## 3 Brazil 1999 37737/172006362 172006362
## 4 Brazil 2000 80488/174504898 174504898
## 5 China 1999 212258/1272915272 1272915272
## 6 China 2000 213766/1280428583 12804285835.4.2.3 Separating by position
It is also possible to separate by position. sep = 2 would split the column between the second and third character/ number etc.
Let’s try that the separation by position. We will keep 4 letters from each country name rather than the whole word, and drop the rest.
separate(table3, country, into = c("country", NA), sep = 4)## # A tibble: 6 x 3
## country year rate
## <chr> <int> <chr>
## 1 Afgh 1999 745/19987071
## 2 Afgh 2000 2666/20595360
## 3 Braz 1999 37737/172006362
## 4 Braz 2000 80488/174504898
## 5 Chin 1999 212258/1272915272
## 6 Chin 2000 213766/12804285835.4.2.4 Separating into more than 2 columns
Special case. Let’s modify table3 a little bit by adding a new column with a value of 200 and unite that column with column rate.
table3_mod <- table3 %>%
mutate(new = 200) %>%
unite(rate, rate, new, sep = "/")Now column rate has 3 sets of numbers, separated by two /. If we define we want to split the data into 2 columns, the first 2 sets of numbers are kept, and the third one is dropped with a warning. The code is still executed but the warning indicates that it might not have gone as planned. Hence, it is always good to check that the data object in the Global Environment looks exactly as what you have intended.
separate(table3_mod, rate, into = c("cases", "population"), sep = "/")## Warning: Expected 2 pieces. Additional pieces discarded in 6 rows [1, 2, 3, 4,
## 5, 6].## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <chr> <chr>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583If we are separating the columns and wanted to keep all sets of numbers, one in each column, we would use an into argument that includes 3 new columns.
separate(table3_mod, rate, into = c("cases", "population", "200"), sep = "/")## # A tibble: 6 x 5
## country year cases population `200`
## <chr> <int> <chr> <chr> <chr>
## 1 Afghanistan 1999 745 19987071 200
## 2 Afghanistan 2000 2666 20595360 200
## 3 Brazil 1999 37737 172006362 200
## 4 Brazil 2000 80488 174504898 200
## 5 China 1999 212258 1272915272 200
## 6 China 2000 213766 1280428583 200
Technically, you would not need a separator argument in the case above, as separate() splits the data whenever it detects a non-alphanumeric character (i.e. a character that isn’t a number or letter). Saying that, the separator argument is needed when your column contains non-alphanumeric characters as part of your cell values.
If the values in population and 200 (separated by a /) were some sort of ID_code, AND we wanted to split the data into 2 columns, after the first / only, we would have to include an extra argument, and set it to "merge".
separate(table3_mod, rate, into = c("cases", "ID_code"), sep = "/", extra = "merge")## # A tibble: 6 x 4
## country year cases ID_code
## <chr> <int> <chr> <chr>
## 1 Afghanistan 1999 745 19987071/200
## 2 Afghanistan 2000 2666 20595360/200
## 3 Brazil 1999 37737 172006362/200
## 4 Brazil 2000 80488 174504898/200
## 5 China 1999 212258 1272915272/200
## 6 China 2000 213766 1280428583/200
There is still a tiny bit off in the tb_cases. Can you spot what it is? * Hint: Think about if you wanted to sum up all the TB cases occurred in the 3 countries within the years 1999, and 2000.
The new columns ID_code and cases are character columns. Whilst this is fine for ID_code, it is less beneficial for cases. If we wanted to sum up the cases column, values would need to be numerical. We could use the converting function as.integer().
table3_mod %>%
separate(rate, into = c("cases", "ID_code"), sep = "/", extra = "merge") %>%
mutate(cases = as.integer(cases))## # A tibble: 6 x 4
## country year cases ID_code
## <chr> <int> <int> <chr>
## 1 Afghanistan 1999 745 19987071/200
## 2 Afghanistan 2000 2666 20595360/200
## 3 Brazil 1999 37737 172006362/200
## 4 Brazil 2000 80488 174504898/200
## 5 China 1999 212258 1272915272/200
## 6 China 2000 213766 1280428583/2005.4.2.5 Converting data types
The slightly quicker and perhaps more elegant solution would be to add another argument convert to separate() and set it to TRUE. This will convert cases to integer and leave ID_code as character due to the non-numerical /.
table3_mod %>%
separate(rate, into = c("cases", "ID_code"), sep = "/", extra = "merge", convert = TRUE)## # A tibble: 6 x 4
## country year cases ID_code
## <chr> <int> <int> <chr>
## 1 Afghanistan 1999 745 19987071/200
## 2 Afghanistan 2000 2666 20595360/200
## 3 Brazil 1999 37737 172006362/200
## 4 Brazil 2000 80488 174504898/200
## 5 China 1999 212258 1272915272/200
## 6 China 2000 213766 1280428583/2005.4.3 unite()
Take a look at table5.
table5## # A tibble: 6 x 4
## country century year rate
## * <chr> <chr> <chr> <chr>
## 1 Afghanistan 19 99 745/19987071
## 2 Afghanistan 20 00 2666/20595360
## 3 Brazil 19 99 37737/172006362
## 4 Brazil 20 00 80488/174504898
## 5 China 19 99 212258/1272915272
## 6 China 20 00 213766/1280428583Here the values in column year are split into century and year which is not very useful. Best to merge the 2 columns together as shown in the image below.

unite() (the inverse of separate) can help to combine the values of century and year into a single column. It is used less frequently than it’s counterpart, but is still a useful function to know.
The function unite() is structured like this:
We would need the data table5, a new column we want to create year, the two columns we would like to combine (century and year), and define the separator as “nothing” since the default is sep = “_“.
unite(table5, year, century, year, sep = "")## # A tibble: 6 x 3
## country year rate
## <chr> <chr> <chr>
## 1 Afghanistan 1999 745/19987071
## 2 Afghanistan 2000 2666/20595360
## 3 Brazil 1999 37737/172006362
## 4 Brazil 2000 80488/174504898
## 5 China 1999 212258/1272915272
## 6 China 2000 213766/1280428583Yay! The data of table5 is in a tidier format now - at least for column year. Ah, but we didn’t save the changes as an object in our Global Environment.
Your turn
Let’s combine the two steps of tidying table5 in a single pipe %>% - separate the rate column into cases and population, and unite the columns year and century into a new column year. Store the new object as tb_cases in the Global Environment. Make sure, the columns cases and population are integer values.
tb_cases <- table5 %>%
separate(rate, into = c("cases", "population"), sep = "/", convert = TRUE) %>%
unite(year, century, year, sep = "")
tb_cases## # A tibble: 6 x 4
## country year cases population
## <chr> <chr> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 1280428583
The year column is still a character column. In this case, it does not matter too much, as 1999 and 2000 are levels of the column year and very unlikely to be involved in any calculations. In case we are dealing with actual numerical value and have the need to convert the data type, we need to use the function as.integer() as unite does not have a convert argument available.
tb_cases_converted <- tb_cases %>%
mutate(year = as.integer(year))
tb_cases_converted## # A tibble: 6 x 4
## country year cases population
## <chr> <int> <int> <int>
## 1 Afghanistan 1999 745 19987071
## 2 Afghanistan 2000 2666 20595360
## 3 Brazil 1999 37737 172006362
## 4 Brazil 2000 80488 174504898
## 5 China 1999 212258 1272915272
## 6 China 2000 213766 12804285835.4.4 additional arguments for unite()
5.4.4.1 Retaining the original column
Similarly to separate(), unite also comes with a remove statement, so if you wanted to keep the original columns before merging them together, you would code
unite(table5, year_combined, century, year, sep = "", remove = FALSE)## # A tibble: 6 x 5
## country year_combined century year rate
## <chr> <chr> <chr> <chr> <chr>
## 1 Afghanistan 1999 19 99 745/19987071
## 2 Afghanistan 2000 20 00 2666/20595360
## 3 Brazil 1999 19 99 37737/172006362
## 4 Brazil 2000 20 00 80488/174504898
## 5 China 1999 19 99 212258/1272915272
## 6 China 2000 20 00 213766/12804285835.4.4.2 Removing missing values before uniting columns
unite() also comes with an na.rm statement that will remove missing values prior to uniting the values of a column (actually, the values have to be a “character” column). Let me modify table5 a wee bit to make that point.
table5_mod <- table5 %>%
mutate(new = rep(c("200", NA), 3))
table5_mod## # A tibble: 6 x 5
## country century year rate new
## <chr> <chr> <chr> <chr> <chr>
## 1 Afghanistan 19 99 745/19987071 200
## 2 Afghanistan 20 00 2666/20595360 <NA>
## 3 Brazil 19 99 37737/172006362 200
## 4 Brazil 20 00 80488/174504898 <NA>
## 5 China 19 99 212258/1272915272 200
## 6 China 20 00 213766/1280428583 <NA>If we wanted to combine the values of rate and new, we would code
unite(table5_mod, rate, rate, new, sep = "/")## # A tibble: 6 x 4
## country century year rate
## <chr> <chr> <chr> <chr>
## 1 Afghanistan 19 99 745/19987071/200
## 2 Afghanistan 20 00 2666/20595360/NA
## 3 Brazil 19 99 37737/172006362/200
## 4 Brazil 20 00 80488/174504898/NA
## 5 China 19 99 212258/1272915272/200
## 6 China 20 00 213766/1280428583/NASee how R adds the missing values onto rate, which we do not want. To avoid that happening, we can use the na.rm statement which removes missing values in columns of data type character before merging the columns.
unite(table5_mod, rate, rate, new, sep = "/", na.rm = TRUE)## # A tibble: 6 x 4
## country century year rate
## <chr> <chr> <chr> <chr>
## 1 Afghanistan 19 99 745/19987071/200
## 2 Afghanistan 20 00 2666/20595360
## 3 Brazil 19 99 37737/172006362/200
## 4 Brazil 20 00 80488/174504898
## 5 China 19 99 212258/1272915272/200
## 6 China 20 00 213766/12804285835.5 Bonus materials: mutate row names from the mtcars
Remember, in lecture 2 when we were talking about the mtcars dataset and how the car types were not in listed as a separate column but rather row names? Let’s just read in the data and have another quick look to jog our memories…
df_mtcars <- mtcars
head(df_mtcars)## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
## Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
## Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
## Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1You can see much better what I’m talking about when you view your df_mtcars.
Now, that we have learnt about the mutate() function, we can attempt to get all of these row names into a new column. All we have to do is combining a function called rownames() with mutate(). We also could use some more of these handy pipes %>% you learnt about last week.
df_mtcars <- mtcars %>%
mutate(Car_type = rownames(mtcars))In this example, we have taken the dataset mtcars, then added a new column called Car_type with mutate(). The column Car_type should then hold all the values that were previously listed as the row names in the original mtcars data.
head(df_mtcars)## mpg cyl disp hp drat wt qsec vs am gear carb Car_type
## 1 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4
## 2 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Mazda RX4 Wag
## 3 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Datsun 710
## 4 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet 4 Drive
## 5 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 Hornet Sportabout
## 6 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 Valiant
We can now look at the data df_mtcars, and see that the new column was added at the end of the dataset. What would we have to do to make Car_type the first column in the dataframe?
# We could use the fuction select()
df_mtcars <- mtcars %>%
mutate(Car_type = rownames(mtcars)) %>%
select(Car_type, mpg:carb)
head(df_mtcars)## Car_type mpg cyl disp hp drat wt qsec vs am gear carb
## 1 Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## 2 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## 3 Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
## 4 Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
## 5 Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
## 6 Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 15.6 Additional information
More information on stuff we covered today and beyond can be found here:
- https://r4ds.had.co.nz “R 4 Data Science” by Garrett Grolemund and Hadley Wickham is the official guidebook for R
- https://suzan.rbind.io/categories/tutorial/ She has some pretty cool tutorials on data wrangling
5.7 Formative Homework
The formative assessment can now be downloaded from moodle. There is no need to submit your answers to moodle.